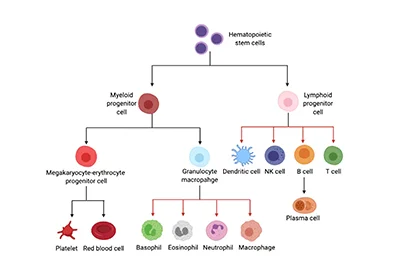
- In-Stock Tumor Cell Lines
- Human Orbital Fibroblasts
- Human Microglia
- Human Pulmonary Alveolar Epithelial Cells
- Human Colonic Fibroblasts
- Human Type II Alveolar Epithelial Cells
- Human Valvular Interstitial Cells
- Human Thyroid Epithelial Cells
- C57BL/6 Mouse Dermal Fibroblasts
- Human Alveolar Macrophages
- Human Dermal Fibroblasts, Adult
- Human Lung Fibroblasts, Adult
- Human Retinal Muller Cells
- Human Articular Chondrocytes
- Human Retinal Pigment Epithelial Cells
- Human Pancreatic Islets of Langerhans Cells
- Human Kidney Podocyte Cells
- Human Renal Proximal Tubule Cells
As one of the most effective tools for gene editing, CRISPR has nowadays been a hotspot technology applied among scientists in the biotechnology industry.
What is CRISPR/Cas9
The clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system is an adaptive immune defense system in bacteria and archaea, which can be used to resist invading viruses and exogenous DNA. CRISPR is a repetitive sequence in the genome of prokaryotes. After bacteria are infected by viruses, they can obtain the DNA fragments of viruses to integrate into the genome and form memories. Cas genome is located near CRISPR. Cas9 protein was encoded by Cas9 gene, and its function is to cut the identified matched genes. When bacteria are invaded again by viruses that have formed memories, bacteria can remove viral genes from their genomes through CRISPR/Cas system. In 2012, Martin et al reconstructed the class II CRISPR/Cas system (CRISPR/Cas9) to make it suitable for genome editing[1].
The mechanism of CRISPR/Cas9 is as follows: CRISPR derived RNA (crRNA) is combined with trans activating RNA (tracrRNA) through base pairing to form tracrRNA/crRNA complex, which is called short guide RNA (sgRNA). The length of sgRNA is generally 20bp. The DNA recognition region of CRISPR is sgRNA, and Cas9 protein is responsible for DNA splicing. sgRNA can guide nuclease (Cas9 protein) to cut double stranded DNA at the target site and form double strand break (DSB)(Figure 1). Cells can repair damaged DNA sequences through homology directed repair (HDR) or non-homologous end joining (NHEJ). HDR mediated precise repair can introduce precise point mutations or insertion mutations from single or double stranded DNA donor templates. NHEJ mediated repair can produce imprecise variable length insertion and/or deletion mutations at DNA DSB sites[2].
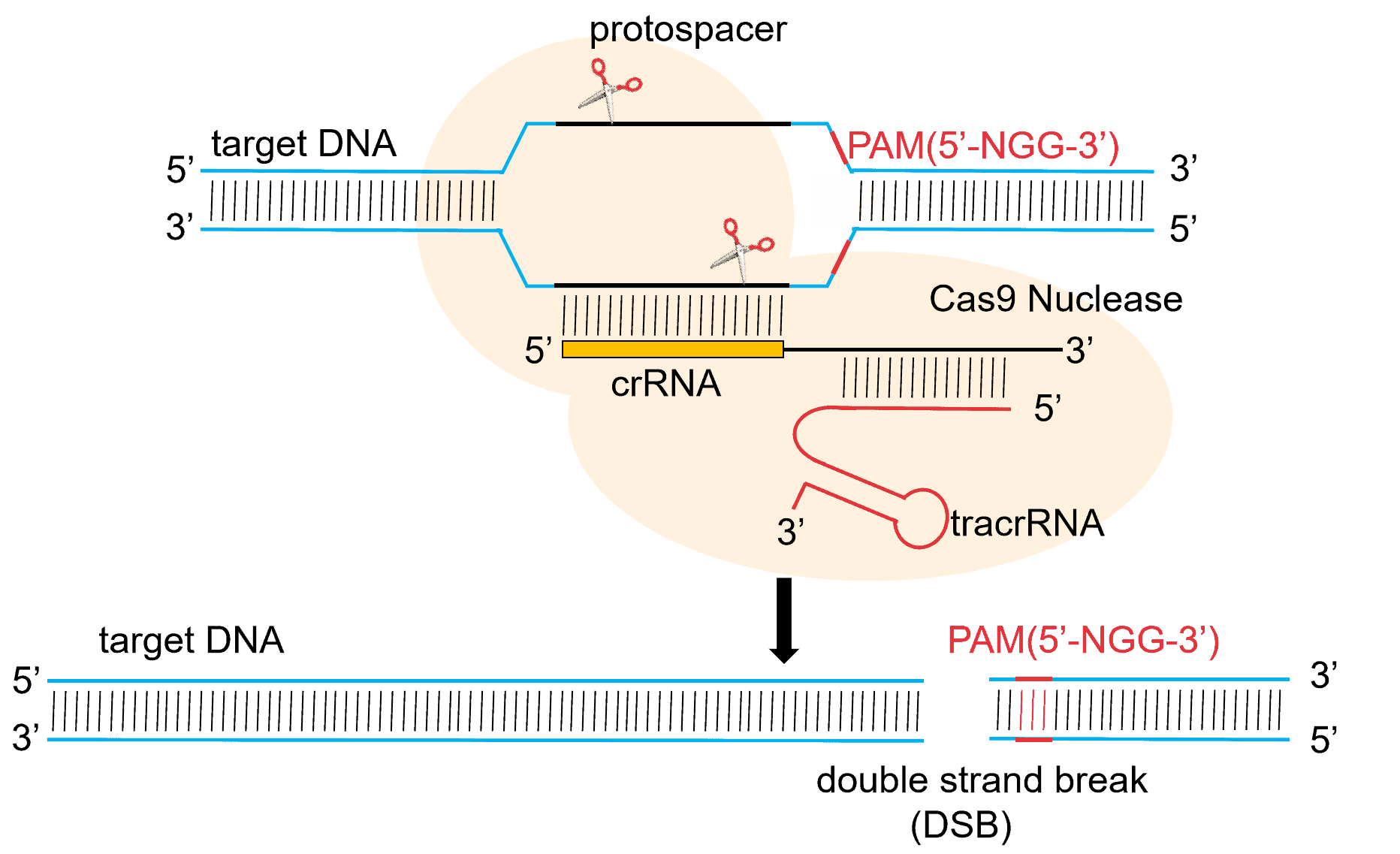
Figure 1. The mechanism of CRISPR/Cas9.
CRISPR/Cas9 is considered as a powerful tool for research and drug discovery, which can change any gene in any cell in a highly targeted manner without introducing exogenous DNA. In 2013, Cong et al. used CRISPR/Cas9 for genome editing in mouse and human cells for the first time [3]. This breakthrough research has written a new chapter in genome editing technology.
Compared with other genome editing technologies, CRISPR/Cas9 gene editing technology has the following advantages: (1) Simple vectors’ construction with accurate targeting; (2) Heritable RNA mediated DNA modification; (3) Highly-efficient gene editing; (4) Simultaneous knockout of multiple genes; (5) No species limitation; (6) Time and money saving[4].
Protocol of gene editing by using CRISPR/Cas9
Highly efficient sgRNA was designed and screened, and the plasmids were transferred into the cells to pick up the single cell clone. Through PCR and sequencing verification, the homozygous cells with gene knockout were successfully obtained.
Determine the target site and design sgRNA (using online tools or self-designed).
Input the nucleotide sequence within 250bp of the target gene. Note that the intron must be avoided. An alternative target site will be given in about 10mins. When manual selection is performed, the 20bp fragment upstream of the target region 5 ‘- NGG (PAM) can be used as the target site. Generally, the target site can be either the sense chain or the antisense chain.
Constructing sgRNA plasmid.
sgRNA was ligated into the sgRNA backbone plasmid, sequenced and aligned. The correctly ligated monoclon was picked and the plasmid was extracted.
Screening high efficiency sgRNA.
Cas9 plasmid and sgRNA plasmid were transferred into cells. After 48-72 hours, cells with fluorescence were sorted by fluorescence activated cell sorting(FACS) or screened by G418. The genome of the screened cells was extracted and the efficiency of sgRNA was detected.
Obtaining knockout cell lines.
High efficiency sgRNA plasmid and Cas9 plasmid were transferred into cells, and the successful transfected monoclonal was screened and genotyped to obtain knockout cell lines.
Application of CRISPR/Cas system in molecular diagnosis
Based on the cleavage characteristics of CRISPR enzyme, molecular diagnosis technology is expected to achieve breakthroughs in two directions: First, to achieve high-sensitivity quantitative detection of multiple pathogens; second, rapid diagnosis and reduced detection time.
Low cost CRISPR/Cas9 based disk diagnostic system for rapid detection of Zika virus.
In 2016, Pardee et al. developed a low-cost rapid diagnostic system based on test paper, which can specifically detect Zika virus, and it may soon be used to screen blood, urine or saliva samples on the spot. The diagnosis module based on CRISPR/Cas9 technology can accurately identify the genetic characteristic sequence of the virus strain[5].
CRISPR-based diagnostic platform: SHERLOCK.
In 2017, Zhang Feng laboratory team found that Cas13 can cleave ssRNA or mRNA (similar to RNA interference) under the guidance of a single crRNA to realize the application of gene knockdown. At the same time, Cas13 also has a nonspecific ssRNA cleavage function. The SHERLOCK V1 method developed by it uses the mixed RNase activity of Cas13 and the quenchable ssRNA reporter group to rapidly detect single DNA or RNA molecules through isothermal amplification steps[6].
In 2019, Zhang Feng laboratory team realized faster and more sensitive detection through characterization and application development of CRISPR enzymology: 1. The lower limit of detection was as low as 2 attomolar; 2. By combining Cas13 with Csm6, an auxiliary enzyme related to CRISPR, the signal sensitivity was increased by 3.5 times; 3. SHERLOCK V2 can detect dengue or Zika virus single stranded RNA and mutations in patient liquid biopsy samples by lateral chromatography, highlighting its potential as a reusable, portable, rapid and quantitative detection platform for nucleic acids[7].
CRISPR-based diagnostic platform: DETECTR.
Professor Jennifer Doudna found that when the Cas12 enzyme family is combined with the target sequence under the guidance of gRNA, it will switch to the active state to cleave other single stranded DNA in the system. This feature of Cas12a can be used in the field of molecular diagnosis to detect tumor genes or specific pathogens. Professor doudna combined the characteristics of Cas12a targeted cleavage of single stranded DNA with recombinant polymerase amplification (RPA) technology. When heated to body temperature, RPA will rapidly increase the copy number of the target DNA, making it easier for Cas12a to find and cut it, and then arbitrarily cut the nearby single stranded DNA, so that the reporter molecule emits fluorescence.
In order to analyze the specificity and sensitivity of the DETECTR method, Professor Doudna tested DNA extracts from 25 human anal swabs, which had previously been analyzed for HPV infection by polymerase chain reaction (PCR). Within 1 hour, detectr accurately identified HPV16 (25/25) and HPV18 (23/25) in patient samples containing a heterogeneous mixture of HPV types, and there was a good correlation between PCR results and detectr fluorescence signals. These results demonstrate a new CRISPR based diagnostic platform, similar to that used for RNA detection using CRISPR/Cas13a, and indicate that detectr can detect any DNA sequence with high sensitivity and specificity in principle[8].
Coronavirus antibody detection: PICASSO technology.
In 2021, scientific researchers have developed a new protein research technology called PICASSO by modifying CRISPR technology. PICASSO technology can be used to detect whether there are antibodies binding to pathogen proteins in the blood of COVID-19 patients[9]. In addition to being a new type of medical diagnosis method, PICASSO is expected to be applied to the experiments of discovering enzyme substrates and protein evolution and design in the future.
New CRISPR tools
In recent years, CRISPR/Cas system has been improved, and many substitutes for CRISPR/Cas system have been discovered, such as Cas9 variants, Cas9 homologs and new Cas proteins other than Cas9. Cas9 variants refer to optimized and improved Cas9 proteins, such as Cas9n[10] and nuclease deficient mutants (dCas9)[11]; Cas9 homologue refers to Cas9 protein found in other strains, for example, Cas9 found in Staphylococcus aureus is called Staphylococcus aureus Cas9 (saCas9)[12,13]; At the same time, many new Cas proteins have been found, such as CRISPR-Cpf1 system[14], CRISPR-C2c1/2/3 system[15], CRISPR-Cas12a system[16] and CRISPR-Cas13 system[17].
Where to Get Knockout Stable Cells Lines for Your Research?
AcceGen is offering a series of gene knockout cell lines developed by the CRISPR/Cas9 system. These cell lines provide you with a convenient means to study gene functions. To get more information, please refer to: Knockout Stable Cell Lines
It is our pleasure to help relative researches to move forward. All the products of AcceGen are strictly comply with international standards. For more detailed information, please visit our product portfolio or contact [email protected].
Reference
[1] Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex
mediates specific DNA cleavage for adaptive immunity in bacteria. PNAS, 2012, 109(39): E2579-2586.
[2] Gaj T, Gersbach CA, Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in Biotechnology, 2013, 31(7):397-405.
[3]Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA,Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121):819-823.
[4]Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9-rapid, efficient and specific choicesfor genome modifications. Journal of Genetics and Genomics, 2013, 40(6):281-289.
[5] Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L, Collins JJ. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell, 2016, 165(5):1255-1266.
[6] Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. 2017, Science, 356:438-442.
[7] Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. 2019, Nat Protoc, 14:2986-3012.
[8] Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. 2018, Science, 360:436-439.
[9] Barber KW, Shrock E, Elledge SJ. CRISPR-based peptide library display and programmable microarray self-assembly for rapid quantitative protein binding assays. Mol Cell, 2021, 81(17):3650-3658.
[10] Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable
dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012,
337(6096):816-821.
[11]Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 2013, 154(2):442-451.
[12]Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal structure of staphylococcus aureus Cas9. Cell, 2015, 162(5):1113-1126.
[13]Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X,Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature, 2015, 520(7546):186-191.
[14]Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 2015, 163(3):759-771.
[15]Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular Cell, 2015, 60(3):385-397.
[16]Teng F, Li J, Cui T, Xu K, Guo L, Gao Q, Feng G, Chen C, Han D, Zhou Q, Li W. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biology, 2019, 20(1):15.
[17]Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13.Nature, 2017, 550(7675):280-284.

Copyright - Unless otherwise stated all contents of this website are AcceGen™ All Rights Reserved – Full details of the use of materials on this site please refer to AcceGen Editorial Policy – Guest Posts are welcome, by submitting a guest post to AcceGen you are agree to the AcceGen Guest Post Agreement – Any concerns please contact [email protected]